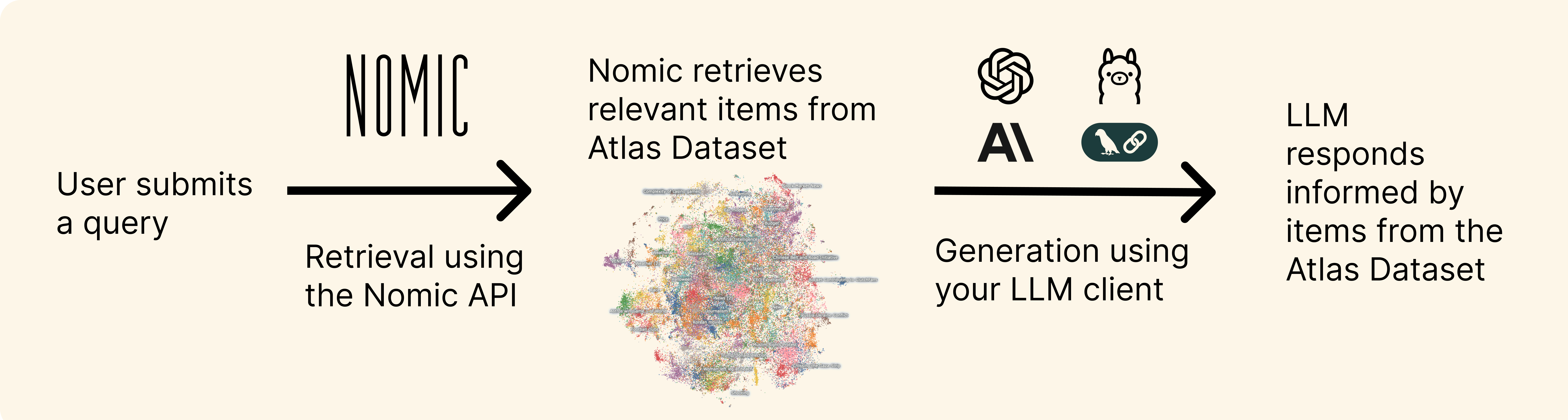
Building RAG Applications with Atlas
Retrieval-augmented generation (RAG) provides large language models (LLMs) additional information retrieved from databases to help answer a user's query.
You can use Atlas as the data layer for retrieval in your RAG application, using the Nomic API to query from an Atlas Dataset via vector search.
This guide shows how to parse and chunk text from a collection of PDFs for a RAG application using Nomic Atlas and the OpenAI Python SDK.
If you prefer, you can follow along with this guide as a Jupyter notebook in our cookbook repo.
Setup
To run the code in this guide, make sure you have docling, nomic, openai, and requests installed to your python environment:
- pip
- uv
pip install docling nomic openai requests
uv add docling nomic openai requests
Then, login to nomic with your Nomic API key. If you don't have a Nomic API key you can create one here.
- terminal
- python
nomic login nk-...
import nomic
nomic.login("nk-...")
Parse PDFs
Let's start with a collection with PDFs and chunk them into snippets to be fetched for retrieval. For this example, we will download and parse PDFs with docling.
from docling.chunking import HybridChunker
from docling.datamodel.base_models import InputFormat
from docling.datamodel.pipeline_options import PdfPipelineOptions
from docling.document_converter import DocumentConverter, PdfFormatOption
pdf_pipeline_options = PdfPipelineOptions(do_ocr=False, do_table_structure=False)
doc_converter = DocumentConverter(
format_options={InputFormat.PDF: PdfFormatOption(
pipeline_options=pdf_pipeline_options
)}
)
chunker = HybridChunker()
# You can replace this with any list of PDFs you want
# 'file' can be a URL or a local filename for a PDF
PDFs = [
{'title': "Attention Is All You Need", 'file': "https://arxiv.org/pdf/1706.03762"},
{'title': "Deep Residual Learning", 'file': "https://arxiv.org/pdf/1512.03385"},
{'title': "BERT", 'file': "https://arxiv.org/pdf/1810.04805"},
{'title': "GPT-3", 'file': "https://arxiv.org/pdf/2005.14165"},
{'title': "Adam Optimizer", 'file': "https://arxiv.org/pdf/1412.6980"},
{'title': "GANs", 'file': "https://arxiv.org/pdf/1406.2661"},
{'title': "U-Net", 'file': "https://arxiv.org/pdf/1505.04597"},
{'title': "DALL-E 2", 'file': "https://arxiv.org/pdf/2204.06125"},
{'title': "Stable Diffusion", 'file': "https://arxiv.org/pdf/2112.10752"}
]
data = []
chunk_id = 0
for pdf in PDFs:
print("Downloading and parsing", pdf['title'])
doc = doc_converter.convert(pdf['file']).document
for chunk in chunker.chunk(dl_doc=doc):
chunk_dict = chunk.model_dump()
filename = chunk_dict['meta']['origin']['filename']
heading = chunk_dict['meta']['headings'][0] if chunk_dict['meta']['headings'] else None
page_num = chunk_dict['meta']['doc_items'][0]['prov'][0]['page_no']
data.append(
{"id": chunk_id, "text": chunk.text, "title": pdf['title'], "filename": filename, "heading": heading, "page_num": page_num}
)
chunk_id += 1
Using docling to parse PDFs provides page number and heading metadata for each document snippet. Additionally, the choice of where one snippet ends and another begins is handled in a smart way by docling: it is good at chunking PDFs according to the actual start and end of sections from the PDFs themselves, as opposed to naive chunking which may start/end chunks awkwardly in the middle of a sentence.
Here we print an item from data to inspect it:
print(data[250])
{'id': 250,
'text': 'We also empirically evaluate the effect of the bias correction terms explained in sections 2 and 3. Discussed in section 5, removal of the bias correction terms results in a version of RMSProp (Tieleman & Hinton, 2012) with momentum. We vary the β 1 and β 2 when training a variational autoencoder (VAE) with the same architecture as in (Kingma & Welling, 2013) with a single hidden layer with 500 hidden units with softplus nonlinearities and a 50-dimensional spherical Gaussian latent variable. We iterated over a broad range of hyper-parameter choices, i.e. β 1 ∈ [0 , 0 . 9] and β 2 ∈ [0 . 99 , 0 . 999 , 0 . 9999] , and log 10 ( α ) ∈ [ -5 , ..., -1] . Values of β 2 close to 1, required for robustness to sparse gradients, results in larger initialization bias; therefore we expect the bias correction term is important in such cases of slow decay, preventing an adverse effect on optimization.\nIn Figure 4, values β 2 close to 1 indeed lead to instabilities in training when no bias correction term was present, especially at first few epochs of the training. The best results were achieved with small values of (1 -β 2 ) and bias correction; this was more apparent towards the end of optimization when gradients tends to become sparser as hidden units specialize to specific patterns. In summary, Adam performed equal or better than RMSProp, regardless of hyper-parameter setting.',
'title': 'Adam Optimizer',
'filename': '1412.6980v9.pdf',
'heading': '6.4 EXPERIMENT: BIAS-CORRECTION TERM',
'page_num': 8}
Create Atlas Dataset
We initialize an AtlasDataset, which we can use to store data and retrieve from it for RAG.
from nomic import AtlasDataset
dataset_identifier = "pdf-data-for-rag" # to create the dataset in the organization connected to your Nomic API key
# dataset_identifier = "<ORG_NAME>/pdf-data-for-rag" # to create the dataset in other organizations you are a member of
atlas_dataset = AtlasDataset(dataset_identifier)
Upload to Atlas
Add the list called data to the AtlasDataset
atlas_dataset.add_data(data)
Create Data Map
Create a data map for this dataset.
We set indexed_field="text" to create embeddings from the text of each PDF chunk, which Atlas uses for visualization and vector search.
atlas_dataset.create_index(
indexed_field="text"
)
Atlas Vector Search API
The Nomic Atlas vector search API returns the k-most semantically similar items from your Atlas Dataset based on a query. You can read more about how to use this endpoint in our API reference here.
This helper function makes an API call to the Nomic Atlas vector search endpoint:
import requests
import os
from nomic import AtlasDataset
def retrieve(query: str, atlas_dataset: AtlasDataset, k: int, fields: list[str]) -> list:
"""Retrieve semantically similar items from an Atlas Dataset based on a query."""
response = requests.post(
'https://api-atlas.nomic.ai/v1/query/topk',
headers={'Authorization': f'Bearer {os.environ.get("NOMIC_API_KEY")}'},
json={
'query': query,
'k': k,
'fields': fields,
'projection_id': atlas_dataset.maps[0].projection_id,
}
)
if response.status_code == 200:
return response.json()['data']
else:
raise ValueError("Invalid API request or incomplete map - if your map hasn't finished building yet, try again once it's ready.")
The parameters for this helper function are:
• query: the text query to search against
• atlas_dataset: the AtlasDataset to retrieve from
• k: number of similar items to return
• fields: which fields/columns from your dataset to return in the response
Let's inspect the output of retrieve on the query "What metrics are mentioned for evaluation?":
query = "What metrics are mentioned for evaluation?"
retrieved_data = retrieve(
query, atlas_dataset, 3, ["title", "heading", "text"]
)
print(retrieved_data)
[{'title': 'Stable Diffusion',
'heading': 'E.3.5 Efficiency Analysis',
'text': 'For efficiency reasons we compute the sample quality metrics plotted in Fig. 6, 17 and 7 based on 5k samples. Therefore, the results might vary from those shown in Tab. 1 and 10. All models have a comparable number of parameters as provided in Tab. 13 and 14. We maximize the learning rates of the individual models such that they still train stably. Therefore, the learning rates slightly vary between different runs cf . Tab. 13 and 14.',
'_similarity': 0.7273091077804565},
{'title': 'GPT-3',
'heading': 'Context → Article:',
'text': "Figure G.11: Formatted dataset example for ARC (Challenge). When predicting, we normalize by the unconditional probability of each answer as described in 2.\nFigure G.13: Formatted dataset example for Winograd. The 'partial' evaluation method we use compares the probability of the completion given a correct and incorrect context.\n53\nFigure G.14: Formatted dataset example for Winogrande. The 'partial' evaluation method we use compares the probability of the completion given a correct and incorrect context.\nContext\n→\nREADING COMPREHENSION ANSWER KEY",
'_similarity': 0.701439619064331},
{'title': 'GPT-3',
'heading': '3 Results',
'text': "Figure 3.1: Smooth scaling of performance with compute. Performance (measured in terms of cross-entropy validation loss) follows a power-law trend with the amount of compute used for training. The power-law behavior observed in [KMH + 20] continues for an additional two orders of magnitude with only small deviations from the predicted curve. For this figure, we exclude embedding parameters from compute and parameter counts.\nTable 3.1: Zero-shot results on PTB language modeling dataset. Many other common language modeling datasets are omitted because they are derived from Wikipedia or other sources which are included in GPT-3's training data. a [RWC + 19]",
'_similarity': 0.6994962692260742}]
RAG with Atlas for Document Retrieval
With a retrieval function for our data map, we can now perform RAG with Atlas as our intermediate data layer we retrieve relevant data from.
We will use GPT4o-mini from OpenAI as our LLM in this example. Make sure you have an OpenAI API key.
import requests
from openai import OpenAI
import os
from nomic import AtlasDataset
client = OpenAI(
# api_key="sk-proj-..." # add your OpenAI API key here, or set it as an environment variable
)
atlas_dataset = AtlasDataset("pdf-data-for-rag")
def retrieve(query: str, atlas_dataset: AtlasDataset, k: int, fields: list[str]) -> list:
"""Retrieve semantically similar items from an Atlas Dataset based on a query."""
response = requests.post(
'https://api-atlas.nomic.ai/v1/query/topk',
headers={'Authorization': f'Bearer {os.environ.get("NOMIC_API_KEY")}'},
json={
'query': query,
'k': k,
'fields': fields,
'projection_id': atlas_dataset.maps[0].projection_id,
}
)
if response.status_code == 200:
return response.json()['data']
else:
raise ValueError("Invalid API request or incomplete map - if your map hasn't finished building yet, try again once it's ready.")
query = "What metrics are mentioned for evaluation?"
retrieved_data = retrieve(
query, atlas_dataset, 3, ["title", "heading", "text"]
)
response = client.chat.completions.create(
model="gpt-4o-mini",
messages=[
{"role": "developer", "content": "You are a helpful assistant. Be specific and cite the context you are given"},
{"role": "user", "content": f"Context:\n{retrieved_data}\n\nQuestion: {query}"}
]
).choices[0].message.content
print(f"Q: {query}\n\nA: {response}")
Q: What metrics are mentioned for evaluation?
A: In the context of the "Stable Diffusion" section, the metrics mentioned for evaluation are "sample quality metrics," which are computed based on 5,000 samples. It indicates that these metrics are relevant for assessing the efficiency of the models. In the "GPT-3" context, evaluation metrics are implied through the descriptions of figures, such as comparing the "probability of the completion given a correct and incorrect context" in the Winograd and Winogrande datasets.
To summarize, the metrics specifically mentioned for evaluation include:
- Sample quality metrics (in Stable Diffusion)
- Probability comparisons in the evaluation methods for the Winograd and Winogrande datasets (in GPT-3).
Jupyter Notebook Guide
You can run this guide as a Jupyter notebook in our cookbook repo.